Our research focuses on the complexity of gene expression through alternative RNA splicing. We aim to gain a deeper understanding of the role of RNA folding in this process and its misregulation in genetic disorders.
RNA splicing plays a profound role in post-transcriptional regulation of genes and contributes significantly to transcriptome and proteome diversity. It is a highly dynamic process that removes non-coding sequences (introns) from the primary transcript and creates mRNA by subsequent ligation of protein-coding segments (exons). Recent findings suggest that almost all human genes undergo alternative splicing by which more than one mRNA transcript is generated from the same mRNA precursor. The accuracy of exon selection is influenced not only by the RNA sequence and interacting proteins but also by the ubiquitous and often complex structures that are formed by RNA. Despite the essential position of RNA splicing in gene expression, the interrelationships between RNA sequence and structure in exon selection remain poorly understood.
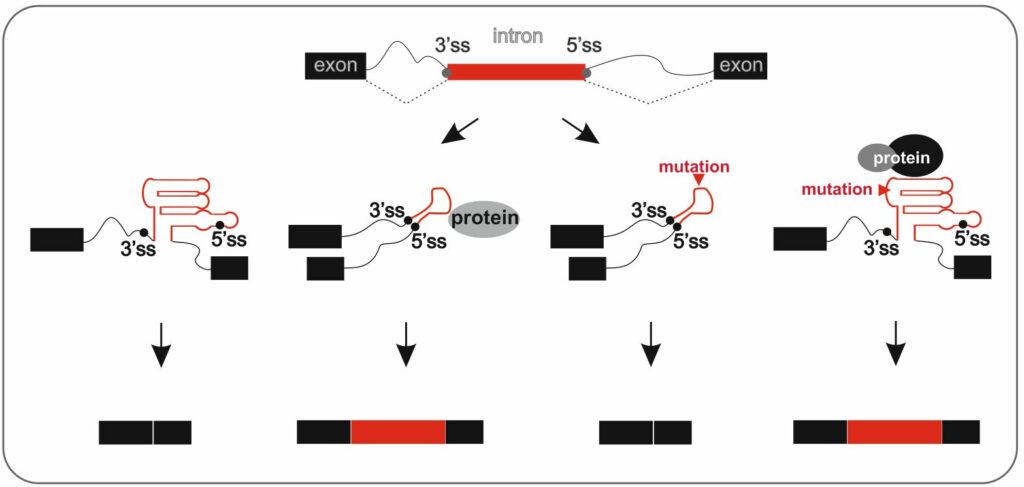
Using a range of state-of-the-art cellular, genetic, biochemical, biophysical, and computational techniques, we focus on:
- understanding the dynamics of conformational changes that occur in pre-mRNA to determine the RNA structural motifs that are critical for exon definition
- the crosstalk between RNA structure and RNA binding proteins in the process of alternative splicing
- the role of transposable elements in the birth of new exons
- characterisation of key RNA conformations of transposable elements induced by evolutionary mutations that underline their exonisation.
Group members
Mgr. Jana Královičová, PhD. – group leader
Ing. Ivana Borovská, PhD. – postdoc
Ing. Lívia Pelegrinová – PhD student
Mgr. Katarína Vondrášková – technician
Funding
RNA structural determinant of Alus exonization
- Program: VEGA
- Duration: 1.1.2022 – 31.12.2025
Selected publications
BOROVSKÁ, Ivana – VOŘECHOVSKÝ, Igor – KRÁLOVIČOVÁ, Jana**. Alu RNA fold links splicing with signal recognition particle proteins. In Nucleic Acids Research, 2023, vol. 51, no. 15, p. 8199-8216. (2022: 14.9 – IF, Q1 – JCR, 8.234 – SJR, Q1 – SJR). ISSN 0305-1048.
ALVAREZ, Maria Elena Vilar – CHIVERS, Martin – BOROVSKÁ, Ivana – MONGER, Steven – GIANNOULATOU, Eleni – KRÁLOVIČOVÁ, Jana – VOŘECHOVSKÝ, Igor**. Transposon clusters as substrates for aberrant splice-site activation. In RNA Biology, 2021, vol. 18, no. 3, p. 354-367. (2020: 4.652 – IF, Q2 – JCR, 2.470 – SJR, Q1 – SJR). ISSN 1547-6286.
KRÁLOVIČOVÁ, Jana* – BOROVSKÁ, Ivana* – PENGELLY, Reuben – LEE, Eunice – ABAFFY, Pavel – ŠINDELKA, Radek – GRUTZNER, Frank – VOŘECHOVSKÝ, Igor**. Restriction of an intron size en route to endothermy. In Nucleic Acids Research, 2021, vol. 49, no. 5, p. 2460-2487. (2020: 16.971 – IF, Q1 – JCR, 9.008 – SJR, Q1 – SJR, karentované – CCC). (2021 – Current Contents). ISSN 0305-1048.
KRÁLOVIČOVÁ, Jana* – BOROVSKÁ, Ivana* – KUBÍČKOVÁ, Monika – LUKAVSKÝ, Peter J. – VOŘECHOVSKÝ, Igor**. Cancer-Associated Substitutions in RNA Recognition Motifs of PUF60 and U2AF65 Reveal Residues Required for Correct Folding and 3 ‘ Splice-Site Selection. In Cancers, 2020, vol. 12, no. 7, art. no. 1865. (2019: 6.126 – IF, Q1 – JCR, 1.938 – SJR, Q1 – SJR). ISSN 2072-6694.
KRÁLOVIČOVÁ, Jana – ŠEVČÍKOVÁ, Ivana – STEJSKALOVÁ, Eva – OBUCA, Mina – HILLER, Michael – STANĚK, David – VOŘECHOVSKÝ, Igor**. PUF60-activated exons uncover altered 3 splice-site selection by germline missense mutations in a single RRM. In Nucleic acids research, 2018, vol. 46, no. 12, p. 6166-6187. (2017: 11.561 – IF, Q1 – JCR, 9.025 – SJR, Q1 – SJR, karentované – CCC). (2018 – Current Contents). ISSN 0305-1048.




